Utilisateur:JeanSolPartre/Noyau
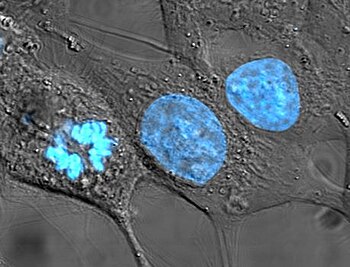
En biologie cellulaire, le noyau est un organite délimité par une membrane, caractéristique des organismes eucaryotes. Les cellules eucaryotes possèdent généralement un seul noyau, mais certains types cellulaires, comme les globules rouges chez les mammifères, n’en ont pas, tandis que d'autres, comme les ostéoclastes, en possèdent de nombreux.
Le noyau peut être observé dans des cellules en interphase. Avec un diamètre variant de 5 à 7 micromètres[1], c’est généralement l’organite le plus visible lorsque l’on observe des cellules au microscope. Pendant le processus de division cellulaire, le noyau disparait temporairement et se reconstitue dans les cellules filles.
Le noyau contient l’essentiel du matériel génétique de la cellule, constitué de plusieurs longues molécules d’ADN associées à une grande variété de protéines, comme les histones, pour former des chromosomes. L'ensemble des gènes localisés sur ces chromosomes constitue le génome nucléaire de la cellule. Le noyau a pour principale fonction de protéger l’intégrité physique de ce génome, et de fournir la machinerie nécessaire à sa réplication et à l’expression de l’information contenue dans les gènes. À ce titre, le noyau peut être considéré comme le centre de contrôle de la cellule.
Le noyau est délimité et isolé du cytoplasme par l'enveloppe nucléaire, une double membrane qui est traversée de nombreux pores permettant le transport nucléo-cytoplasmique de molécules. Les petites molécules et les ions peuvent circuler librement dans les pores nucléaires, mais les macromolécules telles que les ARNm et les protéines doivent devant faire l'objet d'un transport actif reposant sur des transporteurs spécifiques.
L’intérieur du noyau est structuré par la matrice nucléaire, qui comprend elle-même la lamina nucléaire, un réseau interne au noyau qui lui fournit un support mécanique, à l'instar du rôle joué par le cytosquelette dans la cellule. Bien que le noyau ne contienne pas lui-même de sous-compartiments délimités par des membranes, son contenu n'est pas uniforme : il est possible de distinguer un certain nombre d’entités internes au noyau, composées de protéines et d'ARN particuliers, ainsi que de certaines parties des chromosomes. L'exemple le plus connu est le nucléole, qui est principalement impliqué dans l'assemblage des ribosomes, qui sont ensuite exportés vers le cytoplasme où ils accomplissent la traduction des ARNm en protéines.
Histoire modifier
Le noyau est le premier organite à avoir été découvert. C’est à Antonie van Leeuwenhoek (1632–1723), pionnier de la microscopie, que l’on doit le premier dessin connu d'un noyau — qu’il désignait sous le nom de « lumen » — observé dans des globules rouges de saumon.[2] (Contrairement aux mammifères, qui sont un cas particulier, tous les autres vertébrés possèdent des globules rouges nucléés.) Le noyau a également été décrit par Franz Bauer en 1804[3], puis plus en détail en 1831 par le botaniste écossais Robert Brown dans un discours à la Société Linnéenne de Londres. Brown étudiait des orchidées au microscope quand il a observé une zone opaque, qu'il a appelée « aréole » ou « noyau », dans les cellules de la couche externe de la fleur.[4] Il n’a toutefois pas, à l’époque, émis d’hypothèse sur la fonction du noyau.
En 1838, Matthias Schleiden suggère que le noyau joue un rôle dans la production de nouvelle cellules, ce qui l’a amené à proposer le terme de « cytoblaste » (générateur de cellule). Il croyait avoir observé de nouvelles cellules s’assembler autour de ces « cytoblastes ». Franz Meyen était fortement opposé à ce point de vue. Lui qui avait déjà décrit des cellules se multipliant par division, croyait au contraire que la plupart étaient dépourvues de noyau. Il faudra finalement attendre les travaux de Robert Remak (1852) et de Rudolf Virchow (1855) pour que soient jetées les bases d’un nouveau paradigme : les cellules sont générées uniquement par les cellules (« omnis cellula e cellula »), la fonction du noyau demeurant incertaine.[5]
Entre 1876 et 1878, Oscar Hertwig a publié plusieurs études sur la fécondation d’ovules d’oursin, démontrant que le noyau du spermatozoïde pénètre dans l’ovocyte et fusionne avec son noyau. C’était la première fois que l’on suggérait qu’un nouvel individu se développe à partir d’une unique cellule nucléée. Cette idée venait contredire la théorie de la récapitulation formulée par Ernst Haeckel, selon laquelle le développement embryonnaire d’un organisme consistait à passer par une série de stades représentant ses ancêtres phylogénétiques, y compris la génération de la première cellule nucléée à partir d'un « monerula », une masse informe de mucus primordial (Urschleim).
Par conséquent, l’idée que la fécondation nécessite le noyau du spermatozoïde a été débatue pendant un certain temps. Cependant, Hertwig a fini par confirmer ses observations dans d’autres groupes d’animaux, dont les amphibiens et les mollusques. Eduard Strasburger arriva aux mêmes conclusions chez les plantes en 1884. Cela a conduit les scientifiques de l’époque à considérer que le noyau jouait un rôle important dans l’hérédité. En 1873, August Weismann postulait déjà que les cellules germinales paternelle et maternelle participaient de manière équivalente à l’hérédité, mais la fonction du noyau en tant que support de l’information génétique n’a été mise au jour que plus tard, après la découverte de la mitose et la reformulation des lois de la génétique mendélienne au début du XXe siècle, avec la formulation de la théorie chromosomique de l'hérédité.
Structures modifier
The nucleus is the largest cellular organelle in animal cells.[6] In mammalian cells, the average diameter of the nucleus is approximately 6 micrometres (µm), which occupies about 10% of the total cell volume.[7] The viscous liquid within it is called nucleoplasm (or karyolymph), and is similar in composition to the cytosol found outside the nucleus.[8] It appears as a dense, roughly spherical or irregular organelle. The composition by dry weight of the nucleus is approximately: DNA 9%, RNA 1%, Histone Protein 11%, Residual Protein 14%, Acidic Proteins 65%.
In some types of white blood cells specifically most granulocytes the nucleus is lobated and can be present as a bi-lobed, tri-lobed or multi-lobed organelle.
Nuclear envelope and pores modifier
The nuclear envelope, otherwise known as nuclear membrane, consists of two cellular membranes, an inner and an outer membrane, arranged parallel to one another and separated by 10 to 50 nanometres (nm). The nuclear envelope completely encloses the nucleus and separates the cell's genetic material from the surrounding cytoplasm, serving as a barrier to prevent macromolecules from diffusing freely between the nucleoplasm and the cytoplasm.[9] The outer nuclear membrane is continuous with the membrane of the rough endoplasmic reticulum (RER), and is similarly studded with ribosomes.[9] The space between the membranes is called the perinuclear space and is continuous with the RER lumen.
Nuclear pores, which provide aqueous channels through the envelope, are composed of multiple proteins, collectively referred to as nucleoporins. The pores are about 125 million daltons in molecular weight and consist of around 50 (in yeast) to several hundred proteins (in vertebrates).[6] The pores are 100 nm in total diameter; however, the gap through which molecules freely diffuse is only about 9 nm wide, due to the presence of regulatory systems within the center of the pore. This size selectively allows the passage of small water-soluble molecules while preventing larger molecules, such as nucleic acids and larger proteins, from inappropriately entering or exiting the nucleus. These large molecules must be actively transported into the nucleus instead. The nucleus of a typical mammalian cell will have about 3000 to 4000 pores throughout its envelope,[10] each of which contains an eightfold-symmetric ring-shaped structure at a position where the inner and outer membranes fuse.[11] Attached to the ring is a structure called the nuclear basket that extends into the nucleoplasm, and a series of filamentous extensions that reach into the cytoplasm. Both structures serve to mediate binding to nuclear transport proteins.[6]
Most proteins, ribosomal subunits, and some DNAs are transported through the pore complexes in a process mediated by a family of transport factors known as karyopherins. Those karyopherins that mediate movement into the nucleus are also called importins, whereas those that mediate movement out of the nucleus are called exportins. Most karyopherins interact directly with their cargo, although some use adaptor proteins.[12] Steroid hormones such as cortisol and aldosterone, as well as other small lipid-soluble molecules involved in intercellular signaling, can diffuse through the cell membrane and into the cytoplasm, where they bind nuclear receptor proteins that are trafficked into the nucleus. There they serve as transcription factors when bound to their ligand; in the absence of a ligand, many such receptors function as histone deacetylases that repress gene expression.[6]
Nuclear lamina modifier
In animal cells, two networks of intermediate filaments provide the nucleus with mechanical support: The nuclear lamina forms an organized meshwork on the internal face of the envelope, while less organized support is provided on the cytosolic face of the envelope. Both systems provide structural support for the nuclear envelope and anchoring sites for chromosomes and nuclear pores.[7]
The nuclear lamina is composed mostly of lamin proteins. Like all proteins, lamins are synthesized in the cytoplasm and later transported to the nucleus interior, where they are assembled before being incorporated into the existing network of nuclear lamina.[13][14] Lamins found on the cytosolic face of the membrane, such as emerin and nesprin, bind to the cytoskeleton to provide structural support. Lamins are also found inside the nucleoplasm where they form another regular structure, known as the nucleoplasmic veil,[15] that is visible using fluorescence microscopy. The actual function of the veil is not clear, although it is excluded from the nucleolus and is present during interphase.[16] Lamin structures that make up the veil, such as LEM3, bind chromatin and disrupting their structure inhibits transcription of protein-coding genes.[17]
Like the components of other intermediate filaments, the lamin monomer contains an alpha-helical domain used by two monomers to coil around each other, forming a dimer structure called a coiled coil. Two of these dimer structures then join side by side, in an antiparallel arrangement, to form a tetramer called a protofilament. Eight of these protofilaments form a lateral arrangement that is twisted to form a ropelike filament. These filaments can be assembled or disassembled in a dynamic manner, meaning that changes in the length of the filament depend on the competing rates of filament addition and removal.[7]
Mutations in lamin genes leading to defects in filament assembly cause a group of rare genetic disorders known as laminopathies. The most notable laminopathy is the family of diseases known as progeria, which causes the appearance of premature aging in its sufferers. The exact mechanism by which the associated biochemical changes give rise to the aged phenotype is not well understood.[18]
Chromosomes modifier
The cell nucleus contains the majority of the cell's genetic material in the form of multiple linear DNA molecules organized into structures called chromosomes. Each human cell contains roughly two meters of DNA. During most of the cell cycle these are organized in a DNA-protein complex known as chromatin, and during cell division the chromatin can be seen to form the well-defined chromosomes familiar from a karyotype. A small fraction of the cell's genes are located instead in the mitochondria.
There are two types of chromatin. Euchromatin is the less compact DNA form, and contains genes that are frequently expressed by the cell.[19] The other type, heterochromatin, is the more compact form, and contains DNA that is infrequently transcribed. This structure is further categorized into facultative heterochromatin, consisting of genes that are organized as heterochromatin only in certain cell types or at certain stages of development, and constitutive heterochromatin that consists of chromosome structural components such as telomeres and centromeres.[20] During interphase the chromatin organizes itself into discrete individual patches,[21] called chromosome territories.[22] Active genes, which are generally found in the euchromatic region of the chromosome, tend to be located towards the chromosome's territory boundary.[23]
Antibodies to certain types of chromatin organization, in particular, nucleosomes, have been associated with a number of autoimmune diseases, such as systemic lupus erythematosus.[24] These are known as anti-nuclear antibodies (ANA) and have also been observed in concert with multiple sclerosis as part of general immune system dysfunction.[25] As in the case of progeria, the role played by the antibodies in inducing the symptoms of autoimmune diseases is not obvious.
Nucleolus modifier
The nucleolus is a discrete densely stained structure found in the nucleus. It is not surrounded by a membrane, and is sometimes called a suborganelle. It forms around tandem repeats of rDNA, DNA coding for ribosomal RNA (rRNA). These regions are called nucleolar organizer regions (NOR). The main roles of the nucleolus are to synthesize rRNA and assemble ribosomes. The structural cohesion of the nucleolus depends on its activity, as ribosomal assembly in the nucleolus results in the transient association of nucleolar components, facilitating further ribosomal assembly, and hence further association. This model is supported by observations that inactivation of rDNA results in intermingling of nucleolar structures.[26]
In the first step of ribosome assembly, a protein called RNA polymerase I transcribes rDNA, which forms a large pre-rRNA precursor. This is cleaved into the subunits 5.8S, 18S, and 28S rRNA.[27] The transcription, post-transcriptional processing, and assembly of rRNA occurs in the nucleolus, aided by small nucleolar RNA (snoRNA) molecules, some of which are derived from spliced introns from messenger RNAs encoding genes related to ribosomal function. The assembled ribosomal subunits are the largest structures passed through the nuclear pores.[6]
When observed under the electron microscope, the nucleolus can be seen to consist of three distinguishable regions: the innermost fibrillar centers (FCs), surrounded by the dense fibrillar component (DFC), which in turn is bordered by the granular component (GC). Transcription of the rDNA occurs either in the FC or at the FC-DFC boundary, and, therefore, when rDNA transcription in the cell is increased, more FCs are detected. Most of the cleavage and modification of rRNAs occurs in the DFC, while the latter steps involving protein assembly onto the ribosomal subunits occur in the GC.[27]
Other subnuclear bodies modifier
| Structure name | Structure diameter | |
|---|---|---|
| Cajal bodies | 0.2–2.0 µm | [28] |
| Clastosomes | 0.2-0.5 µm | [29] |
| PIKA | 5 µm | [30] |
| PML bodies | 0.2–1.0 µm | [31] |
| Paraspeckles | 0.2–1.0 µm | |
| Speckles | 20–25 nm | [30] |
Besides the nucleolus, the nucleus contains a number of other non-membrane-delineated bodies. These include Cajal bodies, Gemini of coiled bodies, polymorphic interphase karyosomal association (PIKA), promyelocytic leukaemia (PML) bodies, paraspeckles, and splicing speckles. Although little is known about a number of these domains, they are significant in that they show that the nucleoplasm is not a uniform mixture, but rather contains organized functional subdomains.[31]
Other subnuclear structures appear as part of abnormal disease processes. For example, the presence of small intranuclear rods has been reported in some cases of nemaline myopathy. This condition typically results from mutations in actin, and the rods themselves consist of mutant actin as well as other cytoskeletal proteins.[32]
Cajal bodies and gems modifier
A nucleus typically contains between 1 and 10 compact structures called Cajal bodies or coiled bodies (CB), whose diameter measures between 0.2 µm and 2.0 µm depending on the cell type and species.[28] When seen under an electron microscope, they resemble balls of tangled thread[30] and are dense foci of distribution for the protein coilin.[33] CBs are involved in a number of different roles relating to RNA processing, specifically small nucleolar RNA (snoRNA) and small nuclear RNA (snRNA) maturation, and histone mRNA modification.[28]
Similar to Cajal bodies are Gemini of Cajal bodies, or gems, whose name is derived from the Gemini constellation in reference to their close "twin" relationship with CBs. Gems are similar in size and shape to CBs, and in fact are virtually indistinguishable under the microscope.[33] Unlike CBs, gems do not contain small nuclear ribonucleoproteins (snRNPs), but do contain a protein called survival of motor neuron (SMN) whose function relates to snRNP biogenesis. Gems are believed to assist CBs in snRNP biogenesis,[34] though it has also been suggested from microscopy evidence that CBs and gems are different manifestations of the same structure.[33] Later ultrastructural studies have shown gems to be twins of Cajal bodies with the difference being in the coilin component; Cajal bodies are SMN positive and coilin positive, and gems are SMN positive and coilin negative.[35]
RAFA and PTF domains modifier
RAFA domains, or polymorphic interphase karyosomal associations, were first described in microscopy studies in 1991. Their function remains unclear, though they were not thought to be associated with active DNA replication, transcription, or RNA processing.[36] They have been found to often associate with discrete domains defined by dense localization of the transcription factor PTF, which promotes transcription of small nuclear RNA (snRNA).[37]
PML bodies modifier
Promyelocytic leukaemia bodies (PML bodies) are spherical bodies found scattered throughout the nucleoplasm, measuring around 0.1–1.0 µm. They are known by a number of other names, including nuclear domain 10 (ND10), Kremer bodies, and PML oncogenic domains. PML bodies are named after one of their major components, the promyelocytic leukemia protein (PML). They are often seen in the nucleus in association with Cajal bodies and cleavage bodies.[31] PML bodies belong to the nuclear matrix, an ill-defined super-structure of the nucleus proposed to anchor and regulate many nuclear functions, including DNA replication, transcription, or epigenetic silencing.[38] The PML protein is the key organizer of these domains that recruits an ever-growing number of proteins, whose only common known feature to date is their ability to be SUMOylated. Yet, pml-/- mice (which have their PML gene deleted) cannot assemble nuclear bodies, develop normally and live well, demonstrating that PML bodies are dispensable for most basic biological functions.[38]
Splicing speckles modifier
Speckles are subnuclear structures that are enriched in pre-messenger RNA splicing factors and are located in the interchromatin regions of the nucleoplasm of mammalian cells. At the fluorescence-microscope level they appear as irregular, punctate structures, which vary in size and shape, and when examined by electron microscopy they are seen as clusters of interchromatin granules. Speckles are dynamic structures, and both their protein and RNA-protein components can cycle continuously between speckles and other nuclear locations, including active transcription sites. Studies on the composition, structure and behaviour of speckles have provided a model for understanding the functional compartmentalization of the nucleus and the organization of the gene-expression machinery[39] splicing snRNPs[40] and other splicing proteins necessary for pre-mRNA processing. Because of a cell's changing requirements, the composition and location of these bodies changes according to mRNA transcription and regulation via phosphorylation of specific proteins.[41]
The splicing speckles are also known as nuclear speckles (nuclear specks), splicing factor compartments (SF compartments), interchromatin granule clusters (IGCs), B snurposomes. B snurposomes are found in the amphibian oocyte nuclei and in Drosophila melanogaster embryos. B snurposomes appear alone or attached to the Cajal bodies in the electron micrographs of the amphibian nuclei.[42] IGCs function as storage sites for the splicing factors.[43]
Paraspeckles modifier
Discovered by Fox et al. in 2002, paraspeckles are irregularly shaped compartments in the nucleus' interchromatin space.[44] First documented in HeLa cells, where there are generally 10–30 per nucleus, paraspeckles are now known to also exist in all human primary cells, transformed cell lines, and tissue sections.[45] Their name is derived from their distribution in the nucleus; the "para" is short for parallel and the "speckles" refers to the splicing speckles to which they are always in close proximity.
Paraspeckles are dynamic structures that are altered in response to changes in cellular metabolic activity. They are transcription dependent[44] and in the absence of RNA Pol II transcription, the paraspeckle disappears and all of its associated protein components (PSP1, p54nrb, PSP2, CFI(m)68, and PSF) form a crescent shaped perinucleolar cap in the nucleolus. This phenomenon is demonstrated during the cell cycle. In the cell cycle, paraspeckles are present during interphase and during all of mitosis except for telophase. During telophase, when the two daughter nuclei are formed, there is no RNA Pol II transcription so the protein components instead form a perinucleolar cap.[45]
Perichromatin fibrils modifier
Perichromatin fibrils are visible only under electron microscope. They are located next to the transcriptionally active chromatin and are hypothesized to be the sites of active pre-mRNA processing.[43]
Clastosomes modifier
Clastosomes are small nuclear bodies (0.2-0.5 µm) described as having a thick ring-shape due to the peripheral capsule around these bodies.[29] This name is derived from the Greek klastos, broken and soma, body. Clastosomes are not typically present in normal cells, making them hard to detect. They form under high proteolysis conditions within the nucleus and degrade once there is a decrease in activity or if cells are treated with proteasome inhibitors.[29][46] The scarcity of clastosomes in cells indicates that they are not required for proteasome function.[47] Osmotic stress has also been shown to cause the formation of clastosomes.[48] These nuclear bodies contain catalytic and regulatory sub-units of the proteasome and its substrates, indicating that clastosomes are sites for degrading proteins.[47]
Function modifier
The nucleus provides a site for genetic transcription that is segregated from the location of translation in the cytoplasm, allowing levels of gene regulation that are not available to prokaryotes. The main function of the cell nucleus is to control gene expression and mediate the replication of DNA during the cell cycle.
The nucleus is an organelle found in eukaryotic cells. Inside its fully enclosed nuclear membrane, it contains the majority of the cell's genetic material. This material is organized as DNA molecules, along with a variety of proteins, to form chromosomes.
Cell compartmentalization modifier
The nuclear envelope allows the nucleus to control its contents, and separate them from the rest of the cytoplasm where necessary. This is important for controlling processes on either side of the nuclear membrane. In most cases where a cytoplasmic process needs to be restricted, a key participant is removed to the nucleus, where it interacts with transcription factors to downregulate the production of certain enzymes in the pathway. This regulatory mechanism occurs in the case of glycolysis, a cellular pathway for breaking down glucose to produce energy. Hexokinase is an enzyme responsible for the first the step of glycolysis, forming glucose-6-phosphate from glucose. At high concentrations of fructose-6-phosphate, a molecule made later from glucose-6-phosphate, a regulator protein removes hexokinase to the nucleus,[49] where it forms a transcriptional repressor complex with nuclear proteins to reduce the expression of genes involved in glycolysis.[50]
In order to control which genes are being transcribed, the cell separates some transcription factor proteins responsible for regulating gene expression from physical access to the DNA until they are activated by other signaling pathways. This prevents even low levels of inappropriate gene expression. For example, in the case of NF-κB-controlled genes, which are involved in most inflammatory responses, transcription is induced in response to a signal pathway such as that initiated by the signaling molecule TNF-α, binds to a cell membrane receptor, resulting in the recruitment of signalling proteins, and eventually activating the transcription factor NF-κB. A nuclear localisation signal on the NF-κB protein allows it to be transported through the nuclear pore and into the nucleus, where it stimulates the transcription of the target genes.[7]
The compartmentalization allows the cell to prevent translation of unspliced mRNA.[51] Eukaryotic mRNA contains introns that must be removed before being translated to produce functional proteins. The splicing is done inside the nucleus before the mRNA can be accessed by ribosomes for translation. Without the nucleus, ribosomes would translate newly transcribed (unprocessed) mRNA, resulting in malformed and nonfunctional proteins.
Gene expression modifier
Gene expression first involves transcription, in which DNA is used as a template to produce RNA. In the case of genes encoding proteins, that RNA produced from this process is messenger RNA (mRNA), which then needs to be translated by ribosomes to form a protein. As ribosomes are located outside the nucleus, mRNA produced needs to be exported.[52]
Since the nucleus is the site of transcription, it also contains a variety of proteins that either directly mediate transcription or are involved in regulating the process. These proteins include helicases, which unwind the double-stranded DNA molecule to facilitate access to it, RNA polymerases, which bind to the DNA promoter to synthesize the growing RNA molecule, topoisomerases, which change the amount of supercoiling in DNA, helping it wind and unwind, as well as a large variety of transcription factors that regulate expression.[53]
Processing of pre-mRNA modifier
Newly synthesized mRNA molecules are known as primary transcripts or pre-mRNA. They must undergo post-transcriptional modification in the nucleus before being exported to the cytoplasm; mRNA that appears in the cytoplasm without these modifications is degraded rather than used for protein translation. The three main modifications are 5' capping, 3' polyadenylation, and RNA splicing. While in the nucleus, pre-mRNA is associated with a variety of proteins in complexes known as heterogeneous ribonucleoprotein particles (hnRNPs). Addition of the 5' cap occurs co-transcriptionally and is the first step in post-transcriptional modification. The 3' poly-adenine tail is only added after transcription is complete.
RNA splicing, carried out by a complex called the spliceosome, is the process by which introns, or regions of DNA that do not code for protein, are removed from the pre-mRNA and the remaining exons connected to re-form a single continuous molecule. This process normally occurs after 5' capping and 3' polyadenylation but can begin before synthesis is complete in transcripts with many exons.[6] Many pre-mRNAs, including those encoding antibodies, can be spliced in multiple ways to produce different mature mRNAs that encode different protein sequences. This process is known as alternative splicing, and allows production of a large variety of proteins from a limited amount of DNA.
Dynamics and regulation modifier
Nuclear transport modifier
The entry and exit of large molecules from the nucleus is tightly controlled by the nuclear pore complexes. Although small molecules can enter the nucleus without regulation,[54] macromolecules such as RNA and proteins require association karyopherins called importins to enter the nucleus and exportins to exit. "Cargo" proteins that must be translocated from the cytoplasm to the nucleus contain short amino acid sequences known as nuclear localization signals, which are bound by importins, while those transported from the nucleus to the cytoplasm carry nuclear export signals bound by exportins. The ability of importins and exportins to transport their cargo is regulated by GTPases, enzymes that hydrolyze the molecule guanosine triphosphate (GTP) to release energy. The key GTPase in nuclear transport is Ran, which can bind either GTP or GDP (guanosine diphosphate), depending on whether it is located in the nucleus or the cytoplasm. Whereas importins depend on RanGTP to dissociate from their cargo, exportins require RanGTP in order to bind to their cargo.[12]
Nuclear import depends on the importin binding its cargo in the cytoplasm and carrying it through the nuclear pore into the nucleus. Inside the nucleus, RanGTP acts to separate the cargo from the importin, allowing the importin to exit the nucleus and be reused. Nuclear export is similar, as the exportin binds the cargo inside the nucleus in a process facilitated by RanGTP, exits through the nuclear pore, and separates from its cargo in the cytoplasm.
Specialized export proteins exist for translocation of mature mRNA and tRNA to the cytoplasm after post-transcriptional modification is complete. This quality-control mechanism is important due to these molecules' central role in protein translation. Mis-expression of a protein due to incomplete excision of exons or mis-incorporation of amino acids could have negative consequences for the cell; thus, incompletely modified RNA that reaches the cytoplasm is degraded rather than used in translation.[6]
Assembly and disassembly modifier
During its lifetime, a nucleus may be broken down or destroyed, either in the process of cell division or as a consequence of apoptosis (the process of programmed cell death). During these events, the structural components of the nucleus — the envelope and lamina — can be systematically degraded. In most cells, the disassembly of the nuclear envelope marks the end of the prophase of mitosis. However, this disassembly of the nucleus is not a universal feature of mitosis and does not occur in all cells. Some unicellular eukaryotes (e.g., yeasts) undergo so-called closed mitosis, in which the nuclear envelope remains intact. In closed mitosis, the daughter chromosomes migrate to opposite poles of the nucleus, which then divides in two. The cells of higher eukaryotes, however, usually undergo open mitosis, which is characterized by breakdown of the nuclear envelope. The daughter chromosomes then migrate to opposite poles of the mitotic spindle, and new nuclei reassemble around them.
At a certain point during the cell cycle in open mitosis, the cell divides to form two cells. In order for this process to be possible, each of the new daughter cells must have a full set of genes, a process requiring replication of the chromosomes as well as segregation of the separate sets. This occurs by the replicated chromosomes, the sister chromatids, attaching to microtubules, which in turn are attached to different centrosomes. The sister chromatids can then be pulled to separate locations in the cell. In many cells, the centrosome is located in the cytoplasm, outside the nucleus; the microtubules would be unable to attach to the chromatids in the presence of the nuclear envelope.[55] Therefore, the early stages in the cell cycle, beginning in prophase and until around prometaphase, the nuclear membrane is dismantled.[15] Likewise, during the same period, the nuclear lamina is also disassembled, a process regulated by phosphorylation of the lamins by protein kinases such as the CDC2 protein kinase.[56] Towards the end of the cell cycle, the nuclear membrane is reformed, and around the same time, the nuclear lamina are reassembled by dephosphorylating the lamins.[56]
However, in dinoflagellates, the nuclear envelope remains intact, the centrosomes are located in the cytoplasm, and the microtubules come in contact with chromosomes, whose centromeric regions are incorporated into the nuclear envelope (the so-called closed mitosis with extranuclear spindle). In many other protists (e.g., ciliates, sporozoans) and fungi, the centrosomes are intranuclear, and their nuclear envelope also does not disassemble during cell division.
Apoptosis is a controlled process in which the cell's structural components are destroyed, resulting in death of the cell. Changes associated with apoptosis directly affect the nucleus and its contents, for example, in the condensation of chromatin and the disintegration of the nuclear envelope and lamina. The destruction of the lamin networks is controlled by specialized apoptotic proteases called caspases, which cleave the lamin proteins and, thus, degrade the nucleus' structural integrity. Lamin cleavage is sometimes used as a laboratory indicator of caspase activity in assays for early apoptotic activity.[15] Cells that express mutant caspase-resistant lamins are deficient in nuclear changes related to apoptosis, suggesting that lamins play a role in initiating the events that lead to apoptotic degradation of the nucleus.[15] Inhibition of lamin assembly itself is an inducer of apoptosis.[57]
The nuclear envelope acts as a barrier that prevents both DNA and RNA viruses from entering the nucleus. Some viruses require access to proteins inside the nucleus in order to replicate and/or assemble. DNA viruses, such as herpesvirus replicate and assemble in the cell nucleus, and exit by budding through the inner nuclear membrane. This process is accompanied by disassembly of the lamina on the nuclear face of the inner membrane.[15]
modifier
Initially, it has been suspected that immunoglobulins in general and autoantibodies in particular do not enter the nucleus. Now there is a body of evidence that under pathological conditions (e.g. lupus erythematosus) IgG can enter the nucleus.[58]
Nuclei per cell modifier
Most eukaryotic cell types usually have a single nucleus, but some have no nuclei, while others have several. This can result from normal development, as in the maturation of mammalian red blood cells, or from faulty cell division.
Anucleated cells modifier
An anucleated cell contains no nucleus and is, therefore, incapable of dividing to produce daughter cells. The best-known anucleated cell is the mammalian red blood cell, or erythrocyte, which also lacks other organelles such as mitochondria, and serves primarily as a transport vessel to ferry oxygen from the lungs to the body's tissues. Erythrocytes mature through erythropoiesis in the bone marrow, where they lose their nuclei, organelles, and ribosomes. The nucleus is expelled during the process of differentiation from an erythroblast to a reticulocyte, which is the immediate precursor of the mature erythrocyte.[59] The presence of mutagens may induce the release of some immature "micronucleated" erythrocytes into the bloodstream.[60][61] Anucleated cells can also arise from flawed cell division in which one daughter lacks a nucleus and the other has two nuclei.
In flowering plants, this condition occurs in sieve tube elements.
Multinucleated cells modifier
Multinucleated cells contain multiple nuclei. Most acantharean species of protozoa[62] and some fungi in mycorrhizae[63] have naturally multinucleated cells. Other examples include the intestinal parasites in the genus Giardia, which have two nuclei per cell.[64] In humans, skeletal muscle cells, called myocytes and syncytium, become multinucleated during development; the resulting arrangement of nuclei near the periphery of the cells allows maximal intracellular space for myofibrils.[6] Other multinucleate cells in the human are osteoclasts a type of bone cell. Multinucleated and binucleated cells can also be abnormal in humans; for example, cells arising from the fusion of monocytes and macrophages, known as giant multinucleated cells, sometimes accompany inflammation[65] and are also implicated in tumor formation.[66]
A number of dinoflagellates are known to have two nuclei.[67] Unlike other multinucleated cells these nuclei contain two distinct lineages of DNA: one from the dinoflagellate and the other from a symbiotic diatom. The mitochondria and the plastids of the diatom somehow remain functional.
Evolution modifier
As the major defining characteristic of the eukaryotic cell, the nucleus' evolutionary origin has been the subject of much speculation. Four major hypotheses have been proposed to explain the existence of the nucleus, although none have yet earned widespread support.[68]
The first model known as the "syntrophic model" proposes that a symbiotic relationship between the archaea and bacteria created the nucleus-containing eukaryotic cell. (Organisms of the Archaea and Bacteria domain have no cell nucleus.) It is hypothesized that the symbiosis originated when ancient archaea, similar to modern methanogenic archaea, invaded and lived within bacteria similar to modern myxobacteria, eventually forming the early nucleus. This theory is analogous to the accepted theory for the origin of eukaryotic mitochondria and chloroplasts, which are thought to have developed from a similar endosymbiotic relationship between proto-eukaryotes and aerobic bacteria.[69] The archaeal origin of the nucleus is supported by observations that archaea and eukarya have similar genes for certain proteins, including histones. Observations that myxobacteria are motile, can form multicellular complexes, and possess kinases and G proteins similar to eukarya, support a bacterial origin for the eukaryotic cell.[70]
A second model proposes that proto-eukaryotic cells evolved from bacteria without an endosymbiotic stage. This model is based on the existence of modern planctomycetes bacteria that possess a nuclear structure with primitive pores and other compartmentalized membrane structures.[71] A similar proposal states that a eukaryote-like cell, the chronocyte, evolved first and phagocytosed archaea and bacteria to generate the nucleus and the eukaryotic cell.[72]
The most controversial model, known as viral eukaryogenesis, posits that the membrane-bound nucleus, along with other eukaryotic features, originated from the infection of a prokaryote by a virus. The suggestion is based on similarities between eukaryotes and viruses such as linear DNA strands, mRNA capping, and tight binding to proteins (analogizing histones to viral envelopes). One version of the proposal suggests that the nucleus evolved in concert with phagocytosis to form an early cellular "predator".[73] Another variant proposes that eukaryotes originated from early archaea infected by poxviruses, on the basis of observed similarity between the DNA polymerases in modern poxviruses and eukaryotes.[74][75] It has been suggested that the unresolved question of the evolution of sex could be related to the viral eukaryogenesis hypothesis.[76]
A more recent proposal, the exomembrane hypothesis, suggests that the nucleus instead originated from a single ancestral cell that evolved a second exterior cell membrane; the interior membrane enclosing the original cell then became the nuclear membrane and evolved increasingly elaborate pore structures for passage of internally synthesized cellular components such as ribosomal subunits.[77]
Références modifier
- Neil Cambpell, Jane Reece, Lisa Urry, Michael Cain, Steven Wasserman, Peter Minorsky et Robert Jackson, Biologie, Montreuil, France, Pearson, , 9e éd., 1570 p. (ISBN 9782761350655), p. 109
- (la) Antonie van Leeuwenhoek, Opera omnia, seu arcana naturae ope exactissimorum microscopiorum detecta, experimentis variis comprobata, epistolis ad varios illustres viros., Leiden, Pays-Bas, Johan Arnold Langerak,
- (en) Henry Harris, The Birth of the Cell, New Haven, CT, États-Unis, Yale University Press, , 224 p. (ISBN 9780300082951)
- (en) Robert Brown, « On the Organs and Mode of Fecundation of Orchidex and Asclepiadea », Miscellaneous Botanical Works I, vol. 1, , p. 511–514
- (de) Thomas Cremer, Von der Zellenlehre zur Chromosomentheorie : Naturwissenschaftliche Erkenntnis und Theorienwechsel in der frühen Zell- und Vererbungsforschung, Berlin, Allemagne, Springer Verlag, , 1re éd., 384 p. (ISBN 9783642823978)
- (en) Harvey Lodish, Arnold Berk, Paul Matsudaira, Chris A. Kaiser, Monty Krieger, Matthew P. Scott, Lawrence Zipursky et James Darnell, Molecular cell biology, New York, NY, États-Unis, W. H. Freeman and company, , 5e éd., 973 p. (ISBN 9780716743668)
- (en) Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts et Peter Walter, Molecular Biology of the Cell, New York, États-Unis, Garland Science, , 4e éd., 1616 p. (ISBN 9780815332183), chap. 4, p. 191–234
- (en) J. S. Clegg, « Properties and metabolism of the aqueous cytoplasm and its boundaries », American Journal of Physiology, vol. 246, no 2, , R133–R151 (PMID 6364846, DOI 10.1152/ajpregu.1984.246.2.R133)
- (en) Philip L. Paine, Leonard C. Moore et Samuel B. Horowitz, « Nuclear envelope permeability », Nature, vol. 254, no 5496, , p. 109–114 (PMID 1117994, DOI 10.1038/254109a0)
- (en) Rodney Rhoades et Richard Pflanzer, Human physiology, Fort Worth, TX, États-Unis, Saunders College Publishing, , 3e éd., 978 p. (ISBN 9780030051593), chap. 3
- (en) Nataliya Shulga, Nima Mosammaparast, Richard Wozniak et David S. Goldfarb, « Yeast nucleoporins involved in passive nuclear envelope permeability », Journal of Cell Biology, vol. 149, no 5, , p. 1027–1038 (PMID 10831607, DOI 10.1083/jcb.149.5.1027)
- (en) Lucy F. Pemberton L et Bryce M. Paschal, « Mechanisms of receptor-mediated nuclear import and nuclear export », Traffic, vol. 6, no 3, , p. 187–198 (PMID 15702987, DOI 10.1111/j.1600-0854.2005.00270.x)
- (en) Nico Stuurman N, Susanne Heins et Ueli Aebi, « Nuclear lamins: their structure, assembly, and interactions », Journal of Structural Biology, vol. 122, nos 1–2, , p. 42–66 (PMID 9724605, DOI 10.1006/jsbi.1998.3987)
- (en) Anne E. Goldman, Robert D. Moir, Michelle Montag-Lowy, Murray Stewart et Robert D. Goldman, « Pathway of incorporation of microinjected lamin A into the nuclear envelope », Journal of Cell Biology, vol. 119, no 4, , p. 725–735 (PMID 1429833, DOI 10.1083/jcb.119.4.725)
- (en) Robert D. Goldman, Yosef Gruenbaum, Robert D. Moir, Dale K. Shumaker et Timothy P. Spann, « Nuclear lamins: building blocks of nuclear architecture », Genes & Development, vol. 16, no 5, , p. 533–547 (PMID 11877373, DOI 10.1101/gad.960502)
- (en) Robert D. Moir, Miri Yoon, Satya Khuon et Robert D. Goldman, « Nuclear Lamins A and B1: Different Pathways of Assembly during Nuclear Envelope Formation in Living Cells », Journal of Cell Biology, vol. 151, no 6, , p. 1155–1168 (PMID 11121432, DOI 10.1083/jcb.151.6.1155)
- (en) Timothy P. Spann, Anne E. Goldman, Chen Wang, Sui Huang et Robert D. Goldman, « Alteration of nuclear lamin organization inhibits RNA polymerase II–dependent transcription », Journal of Cell Biology, vol. 156, no 4, , p. 603–608 (PMID 11854306, DOI 10.1083/jcb.200112047)
- (en) Leslie C. Mounkes et Colin L. Stewart, « Aging and nuclear organization: lamins and progeria », Current Opinion in Cell Biology, vol. 16, no 3, , p. 322–327 (PMID 15145358, DOI 10.1016/j.ceb.2004.03.009)
- (en) Ann E. Ehrenhofer-Murray, « Chromatin dynamics at DNA replication, transcription and repair », European Journal of Biochemistry, vol. 271, no 12, , p. 2335–2349 (PMID 15182349, DOI 10.1111/j.1432-1033.2004.04162.x)
- (en) Sergei A. Grigoryev, Yaroslava A. Bulynko et Evegnya Y. Popova, « The end adjusts the means: heterochromatin remodelling during terminal cell differentiation », Chromosome Research, vol. 14, no 1, , p. 53–69 (PMID 16506096, DOI 10.1007/s10577-005-1021-6)
- (en) Margit Schardin, T. Cremer, H. D. Hager et M. Lang, « Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories », Human Genetics, vol. 71, no 4, , p. 281–287 (PMID 2416668, DOI 10.1007/BF00388452)
- (en) Angus I. Lamond et William C. Earnshaw, « Structure and Function in the Nucleus », Science, vol. 280, no 5363, , p. 547–553 (PMID 9554838, DOI 10.1126/science.280.5363.547)
- (en) Anette Kurz, Stefan Lampel, Jeremy E. Nickolenko, Joachim Bradl, Axel Benner, Rebekka M. Zirbel, Thomas Cremer et Peter Lichter, « Active and inactive genes localize preferentially in the periphery of chromosome territories », Journal of Cell Biology, vol. 135, no 5, , p. 1195–1205 (PMID 8947544, DOI 10.1083/jcb.135.5.1195)
- (en) Naomi F. Rothfield et B. David Stollar, « The Relation of Immunoglobulin Class, Pattern of Antinuclear Antibody, and Complement-Fixing Antibodies to DNA in Sera from Patients with Systemic Lupus Erythematosus », Journal of Clinical Investigation, vol. 46, no 11, , p. 1785–1794 (PMID 4168731, DOI 10.1172/JCI105669)
- (en) Susan Barned, Andrew D. Goodman et David H. Mattson, « Frequency of anti-nuclear antibodies in multiple sclerosis », Neurology, vol. 45, no 2, , p. 384–385 (PMID 7854544, DOI 10.1212/WNL.45.2.384)
- (en) Danièle Hernandez-Verdun, « Nucleolus: from structure to dynamics », Histochemistry and Cell Biology, vol. 125, nos 1–2, , p. 127–137 (PMID 16328431, DOI 10.1007/s00418-005-0046-4)
- (en) Angus I. Lamond et Judith E. Sleeman, « Nuclear substructure and dynamics », Current Biology, vol. 13, no 21, , R825–R828 (PMID 14588256, DOI 10.1016/j.cub.2003.10.012)
- (en) Mario Cioce et Angus I. Lamond, « Cajal bodies: a long history of discovery », Annual Review of Cell and Developmental Biology, vol. 21, , p. 105–131 (PMID 16212489, DOI 10.1146/annurev.cellbio.20.010403.103738)
- (en) Miguel Lafarga, Miguel Lafarga, Maria Teresa Berciano, Emma Pena, Isabel Mayo, Jose G. Castaño, Dirk Bohmann, João Pedro Rodrigues, João Paulo Tavanez et Maria Carmo-Fonseca, « Clastosome: A Subtype of Nuclear Body Enriched in 19S and 20S Proteasomes, Ubiquitin, and Protein Substrates of Proteasome », Molecular Biology of the Cell, vol. 13, no 8, , p. 2771–2782 (PMID 12181345, DOI 10.1091/mbc.e02-03-0122)
- (en) Thomas D. Pollard et William C. Earnshaw, Cell biology, Philadelphie, PA, États-Unis, Saunders, , 813 p. (ISBN 9780721633602)
- (en) Miroslav Dundr et Tom Misteli, « Functional architecture in the cell nucleus », Biochemical Journal, vol. 356, no 2, , p. 297–310 (PMID 11368755, DOI 10.1042/bj3560297)
- (en) Hans H. Goebel et Irene Warlo, « Nemaline myopathy with intranuclear rods—intranuclear rod myopathy », Neuromuscular Disorders, vol. 7, no 1, , p. 13–19 (PMID 9132135, DOI 10.1016/S0960-8966(96)00404-X)
- (en) A. Gregory Matera et Mark R. Frey, « Coiled Bodies and Gems: Janus or Gemini? », American Journal of Human Genetics, vol. 63, no 2, , p. 317–321 (PMID 9683623, DOI 10.1086/301992)
- (en) A. Gregory Matera, « Of coiled bodies, gems, and salmon. », Journal of Cellular Biochemistry, vol. 70, no 2, , p. 181–192 (PMID 9671224, DOI 10.1002/(SICI)1097-4644(19980801)70:2<181::AID-JCB4>3.0.CO;2-K)
- (en) William S. Saunders, Carol A. Cooke et William C. Earnshaw, « Compartmentalization within the nucleus: discovery of a novel subnuclear region. », Journal of Cell Biology, vol. 115, no 4, , p. 919–931 (PMID 1955462, DOI 10.1083/jcb.115.4.919)
- (en) Ana Pombo, Paula Cuello, Wouter Schul, Jong-Bok Yoon, Robert G. Roeder, Peter R. Cook et Shona Murphy, « Regional and temporal specialization in the nucleus: a transcriptionally active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle », EMBO Journal, vol. 17, no 6, , p. 1768–1778 (PMID 9501098, DOI 10.1093/emboj/17.6.1768)
- (en) Valérie Lallemand-Breitenbach et Hughes de Thé, « PML Nuclear Bodies », Cold Spring Harbor Perspectives in Biology, vol. 2, no 5, , a000661 (PMID 20452955, DOI 10.1101/cshperspect.a000661)
- (en) Angus I. Lamond et David L. Spector, « Nuclear speckles: a model for nuclear organelles », Nature Reviews Molecular Cell Biology, vol. 4, no 8, , p. 605–12 (PMID 12923522, DOI 10.1038/nrm1172)
- (en) Kaushlendra Tripathi et Veena K. Parnaik, « Differential dynamics of splicing factor SC35 during the cell cycle », Journal of Biosciences, vol. 33, no 3, , p. 345–354 (PMID 19005234, DOI 10.1007/s12038-008-0054-3)
- (en) Korie E. Handwerger et Joseph G. Gall, « Subnuclear organelles: new insights into form and function », Trends in Cell Biology, vol. 16, no 1, , p. 19–26 (PMID 16325406, DOI 10.1016/j.tcb.2005.11.005)
- (en) Joseph G. Gall, Michel Bellini, Zheng'an Wu et Christine Murphy, « Assembly of the Nuclear Transcription and Processing Machinery: Cajal Bodies (Coiled Bodies) and Transcriptosomes », Molecular Biology of the Cell, vol. 10, no 12, , p. 4385–4402 (PMID 10588665, DOI 10.1091/mbc.10.12.4385)
- (en) A. Gregory Matera, Rebecca M. Terns et Michael P. Terns, « Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs », Nature Reviews Molecular Cell Biology, vol. 8, no 3, , p. 209–220 (PMID 17318225, DOI 10.1038/nrm2124)
- (en) Archa H. Fox, Yun W. Lam, Anthony K. L. Leung, Carol E. Lyon, Jens Andersen, Matthias Mann et Angus I. Lamond, « Paraspeckles: A Novel Nuclear Domain », Current Biology, vol. 12, no 1, , p. 13–25 (PMID 11790299, DOI 10.1016/S0960-9822(01)00632-7)
- (en) Archa H. Fox, Charles S. Bond et Angus i. Lamond, « P54nrb Forms a Heterodimer with PSP1 That Localizes to Paraspeckles in an RNA-dependent Manner », Molecular Biology of the Cell, vol. 16, no 11, , p. 5304–5315 (PMID 16148043, DOI 10.1091/mbc.e05-06-0587)
- (en) Xiao-Ni Kong, He-Xin Yan, Lei Chen, Li-Wei Dong, Wen Yang, Qiong Liu, Le-Xing Yu, Dan-Dan Huang, Shu-Qin Liu, Hui Liu, Meng-Chao Wu et Hong-Yang Wang, « LPS-induced down-regulation of signal regulatory protein α contributes to innate immune activation in macrophages », Journal of Experimental Medicine, vol. 204, no 11, , p. 2719–2731 (PMID 17954568, DOI 10.1084/jem.20062611)
- (en) Maria Carmo-Fonseca, Maria T. Berciano et Miguel Lafarga, « Orphan Nuclear Bodies », Cold Spring Harbor Perspectives in Biology, vol. 2, no 9, , a000703 (PMID 20610547, DOI 10.1101/cshperspect.a000703)
- (en) Katherine M. Sampuda, Mason Riley et Lynn Boyd, « Stress induced nuclear granules form in response to accumulation of misfolded proteins in Caenorhabditis elegans », BMC Cell Biology, vol. 18, , p. 18 (PMID 28424053, DOI 10.1186/s12860-017-0136-x)
- (en) Albert L. Lehninger, David L. Nelson et Michael M. Cox, Principles of biochemistry, New York, NY, États-Unis, Worth Publishers, , 3e éd., 1152 p. (ISBN 9781572591530)
- (en) F. Moreno, D. Ahuatzi, A. Riera, C. A. Palomino et P. Herrero, « Glucose sensing through the Hxk2-dependent signalling pathway. », Biochemical Society Transactions, vol. 33, no 1, , p. 265–268 (PMID 15667322, DOI 10.1042/BST0330265)
- (en) Dirk Görlich et Ulrike Kutay, « Transport between the cell nucleus and the cytoplasm », Annual Review of Cell and Developmental Biology, vol. 15, no 1, , p. 607–660 (PMID 10611974, DOI 10.1146/annurev.cellbio.15.1.607)
- (en) Kund H. Nierhaus et Daniel N. Wilson, Protein Synthesis and Ribosome Structure: Translating the Genome, Weinheim, Allemagne, Wiley-VCH, , 9e éd., 579 p. (ISBN 9783527306381)
- {{Ouvrage | langue = en | nom1 = Nicolini | prénom1 = Claudio | titre = Genome Structure and Function | sous-titre = From Chromosomes Characterization to Genes Technology | année = 1997 | éditeur = Springer Netherlands | lieu = Dordrecht, Pays-Bas | pages totales = 331 | isbn = 9780792345657
- (en) James D. Watson, Tania A. Baker, Stephen P. Bell, Alexander Gann, Michael Levine et Richard Losick, Molecular biology of the gene, Montreuil, France, Pearson, , 5e éd., 732 p. (ISBN 9780805346350), chap. 9−10
- (en) Jennifer Lippincott-Schwartz, « Cell biology: Ripping up the nuclear envelope », Nature, vol. 416, no 6876, , p. 31–32 (PMID 11882878, DOI 10.1038/416031a)
- (en) Teni Boulikas, « Phosphorylation of transcription factors and control of the cell cycle », Critical Reviews in Eukaryotic Gene Expression, vol. 5, no 1, , p. 1–77 (PMID 7549180)
- (en) Rikke L. Steen et Philippe Collas, « Mistargeting of B-type lamins at the end of mitosis: implications on cell survival and regulation of lamins A/C expression », Journal of Cell Biology, vol. 153, no 3, , p. 621–626 (PMID 11331311, DOI 10.1083/jcb.153.3.621)
- (en) Ingrid Böhm, « IgG deposits can be detected in cell nuclei of patients with both lupus erythematosus and malignancy », Clinical Rheumatology, vol. 26, no 11, , p. 1877–1882 (PMID 17364135, DOI 10.1007/s10067-007-0597-y)
- (en) E. Skutelsky et D. Danon, « Comparative study of nuclear expulsion from the late erythroblast and cytokinesis », Experimental Cell Research, vol. 60, no 3, , p. 625–635 (PMID 5422968, DOI 10.1016/0014-4827(70)90536-7)
- (en) Dorothea K. Torous, Stephen D. Dertinger, Nikki E. Hall et Carol R. Tometsko, « Enumeration of micronucleated reticulocytes in rat peripheral blood: a flow cytometric study », Mutation Research, vol. 465, nos 1–2, , p. 91–99 (PMID 10708974, DOI 10.1016/S1383-5718(99)00216-8)
- (en) K.-J. Hutter et M. Stöhr, « Rapid detection of mutagen induced micronucleated erythrocytes by flow cytometry », Histochemistry, vol. 75, no 3, , p. 353–362 (PMID 7141888, DOI 10.1007/bf00496738)
- (en) Linda Amaral Zettler, Mitchell L. Sogin et David A. Caron, « Phylogenetic relationships between the Acantharea and the Polycystinea: A molecular perspective on Haeckel's Radiolaria », Proceedings of the National Academy of Sciences of the United States of America, vol. 94, no 21, , p. 11411–11416 (PMID 9326623, DOI 10.1073/pnas.94.21.11411)
- (en) Thomas R. Horton, « The number of nuclei in basidiospores of 63 species of ectomycorrhizal Homobasidiomycetes », Mycologia, vol. 98, no 2, , p. 233–238 (PMID 16894968, DOI 10.1080/15572536.2006.11832695)
- (en) Rodney D. Adam, « The biology of Giardia spp. », Microbiological Reviews, vol. 55, no 4, , p. 706–732 (PMID 1779932, PMCID 372844)
- (en) Alison McInnes et Donna M. Rennick, « Interleukin 4 induces cultured monocytes/macrophages to form giant multinucleated cells », Journal of Experimental Medicine, vol. 167, no 2, , p. 598–611 (PMID 3258008, DOI 10.1084/jem.167.2.598)
- (en) Steven R. Goldring, Merrilee S. Roelke, Kathleen K. Petrison et Atul K. Bhan, « Human giant cell tumors of bone identification and characterization of cell types », Journal of Clinical Investigation, vol. 79, no 2, , p. 483–491 (PMID 3027126, DOI 10.1172/JCI112838)
- (en) Behzad Imanian, Jean-François Pombert, Richard G. Dorrell, Fabien Burki et Patrick J. Keeling, « Tertiary endosymbiosis in two dinotoms has generated little change in the mitochondrial genomes of their dinoflagellate hosts and diatom endosymbionts », PLoS One, vol. 7, no 8, , e43763 (PMID 22916303, DOI 10.1371/journal.pone.0043763)
- (en) Elizabeth Pennisi, « Evolutionary biology. The birth of the nucleus », Science, vol. 305, no 5685, , p. 766–768 (PMID 15297641, DOI 10.1126/science.305.5685.766)
- (en) Lynn Margulis, Symbiosis in cell evolution : Life and its environment on the early Earth, San Francisco, CA, États-Unis, W. H. Freeman and company, , 419 p. (ISBN 9780716712558), p. 206–227
- (en) Purificación López-García et David Moreira, « Selective forces for the origin of the eukaryotic nucleus », BioEssays, vol. 28, no 5, , p. 525–533 (PMID 16615090, DOI 10.1002/bies.20413)
- (en) John A. Fuerst, « Intracellular compartmentation in planctomycetes », Annual Review of Microbiology, vol. 59, , p. 299–328 (PMID 15910279, DOI 10.1146/annurev.micro.59.030804.121258)
- (en) Hyman Hartman et Alexei Fedorov, « The origin of the eukaryotic cell: a genomic investigation », Proceedings of the National Academy of Sciences of the United States of America, vol. 99, no 3, , p. 1420–1425 (PMID 11805300, DOI 10.1073/pnas.032658599)
- (en) Philip J. L. Bell, « Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? », Journal of Molecular Evolution, vol. 53, no 3, , p. 251–256 (PMID 11523012, DOI 10.1007/s002390010215)
- (en) Masaharu Takemura, « Poxviruses and the origin of the eukaryotic nucleus », Journal of Molecular Evolution, vol. 52, no 5, , p. 419–425 (PMID 11443345, DOI 10.1007/s002390010171)
- (en) Luis P. Villarreal et Victor R. DeFilippis, « A hypothesis for DNA viruses as the origin of eukaryotic replication proteins », Journal of Virology, vol. 74, no 15, , p. 7079–7084 (PMID 10888648, DOI 10.1128/JVI.74.15.7079-7084.2000)
- (en) Philip J. L. Bell, « Sex and the eukaryotic cell cycle is consistent with a viral ancestry for the eukaryotic nucleus », Journal of Theoretical Biology, vol. 243, no 1, , p. 54–63 (PMID 16846615, DOI 10.1016/j.jtbi.2006.05.015)
- (en) Albert D. G. de Roos, « The origin of the eukaryotic cell based on conservation of existing interfaces », Artificial Life, vol. 12, no 4, , p. 513–523 (PMID 16953783, DOI 10.1162/artl.2006.12.4.513)